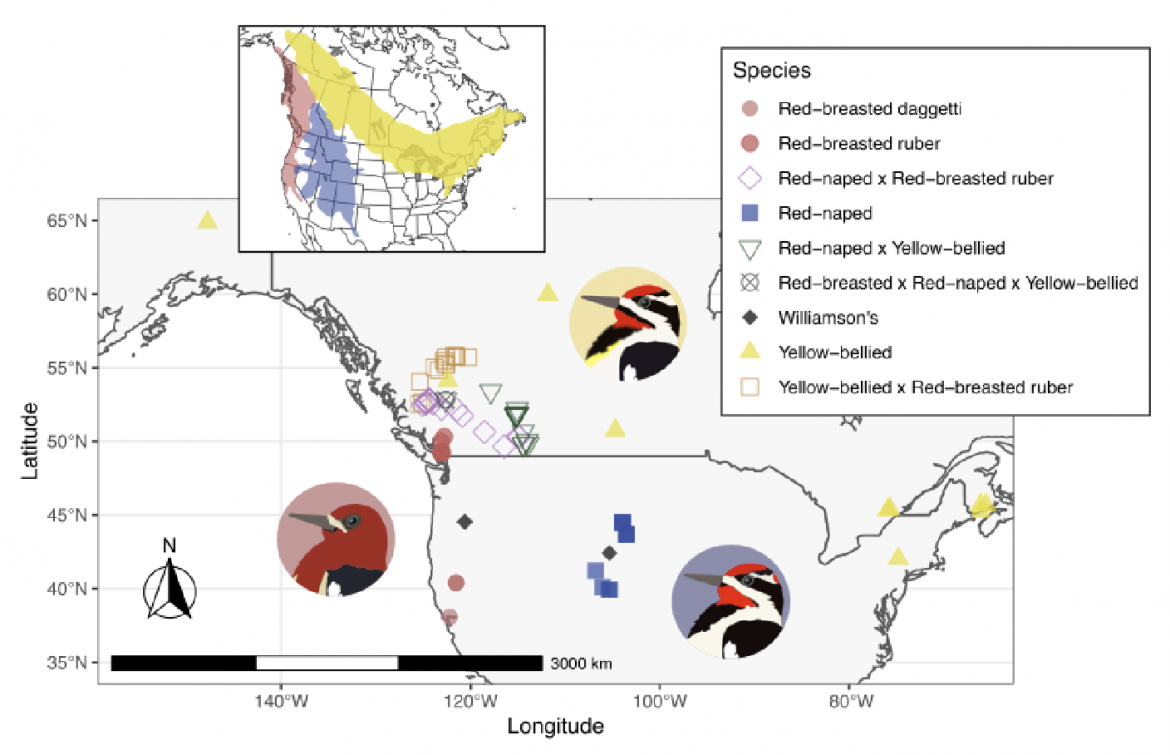
Figure 1 Sampling map of birds included in the WGS sequencing dataset (see Supplemental
Abstract
Speciation occurs when gene pools differentiate between populations, but that differentiation is heterogeneous across the genome. Understanding what parts of the genome are more prone to differentiation is a key mission in evolutionary biology because it can identify genomic regions and evolutionary processes central to the speciation process. Here, we study genomic variation among three closely related species of North American woodpecker: red-breasted, red-naped, and yellow-bellied sapsuckers. We construct a new sapsucker reference genome and use whole genome resequencing to measure genetic variation among these species and to quantify how the level of differentiation between these forms varies across the genome. We find that regions of high relative differentiation between species (FST) tend to have low absolute differentiation between species (πB), indicating that regions of high relative differentiation often have more recent between-population coalescence times than regions of low relative differentiation do. Most of the high-FST genomic windows are found on the Z chromosome, indicating this sex chromosome is particularly important in sapsucker speciation. We show that LD is high across the Z chromosome and that each species tends to have a unique Z haploblock, a pattern suggestive of chromosomal inversions. These results are consistent with a model of speciation in which selective sweeps of globally advantageous variants spread among partly differentiated populations, followed by differential local adaptation of those same genomic regions. We propose that sapsucker speciation may occur primarily via this process occurring on the Z chromosomes, followed by epistatic interactions between Z haploblocks.